Accessing the data🗃️¶
Please cite this publication whenewer referring to the dataset Thummerer A, et al. SynthRAD2025 Grand Challenge dataset: Generating synthetic CTs for radiotherapy from head to abdomen. Med Phys. 2025 Jul;52(7):e17981. https://doi.org/10.1002/mp.17981.
A cohort of 890 (Task 1) and 1472 (Task 2) cases (considering
all the challenge phases) has been prepared for participants. The
training data will be released on 1/03/2025, along with a description of
the imaging parameters and other relevant data.
The dataset is accessible through https://doi.org/10.5281/zenodo.14918089 under the SynthRAD2025 collection. A paper describing the dataset is published in Medical Physics https://doi.org/10.1002/mp.17981.
Data location and release dates¶
| Subset | Files | Release Date | Link |
|---|---|---|---|
| Training | Input, CT, Mask | 01/03/2025 | 10.5281/zenodo.14918213 |
| Training Center D ⚠️ | Input, CT, Mask | 01/03/2025 | Download, License |
| Validation | Input, Mask | 01/06/2025 | 10.5281/zenodo.14918504 |
| Validation Center D ⚠️ | Input, Mask | 01/06/2025 | Download, License |
| Validation | CT, Deformed CT | 01/03/2030 | 10.5281/zenodo.14918605 |
| Testing | Input, CT, Deformed CT, Mask | 01/03/2030 | 10.5281/zenodo.14918722 |
⚠️: Data from center D is provided with a limited license which permits it's use only for the duration of the challenge and remains valid only while the challenge is active (Limited Use License Center D). By downloading Center D's data, participants agree to these terms. Once the challenge ends, access to the data ends, the download link will be deactivated, and all downloaded data must be deleted.
Data structure
After unzipping, each Task is organized according to the following folder structure:
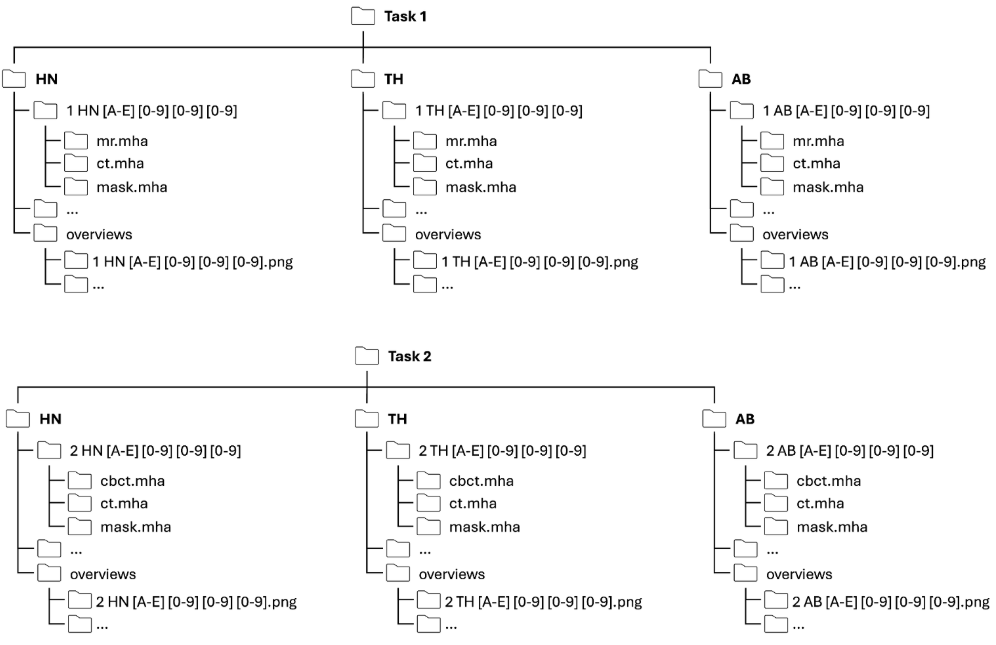
Each patient folder has a unique name that contains information about the task, anatomy and a patient ID. The naming follows the convention below:
[Task] [Anatomy] [CenterID] [PatientID]
For example: 1ABC001 (Task 1, Abdomen, Center C, Patient 001)
For the training dataset each patient folder contains three files:
-
ct.mha: CT image
-
mr.mha or cbct.mha (depending on the task): CBCT/MRI
-
mask.mha: image containing a binary mask of the dilated patient outline
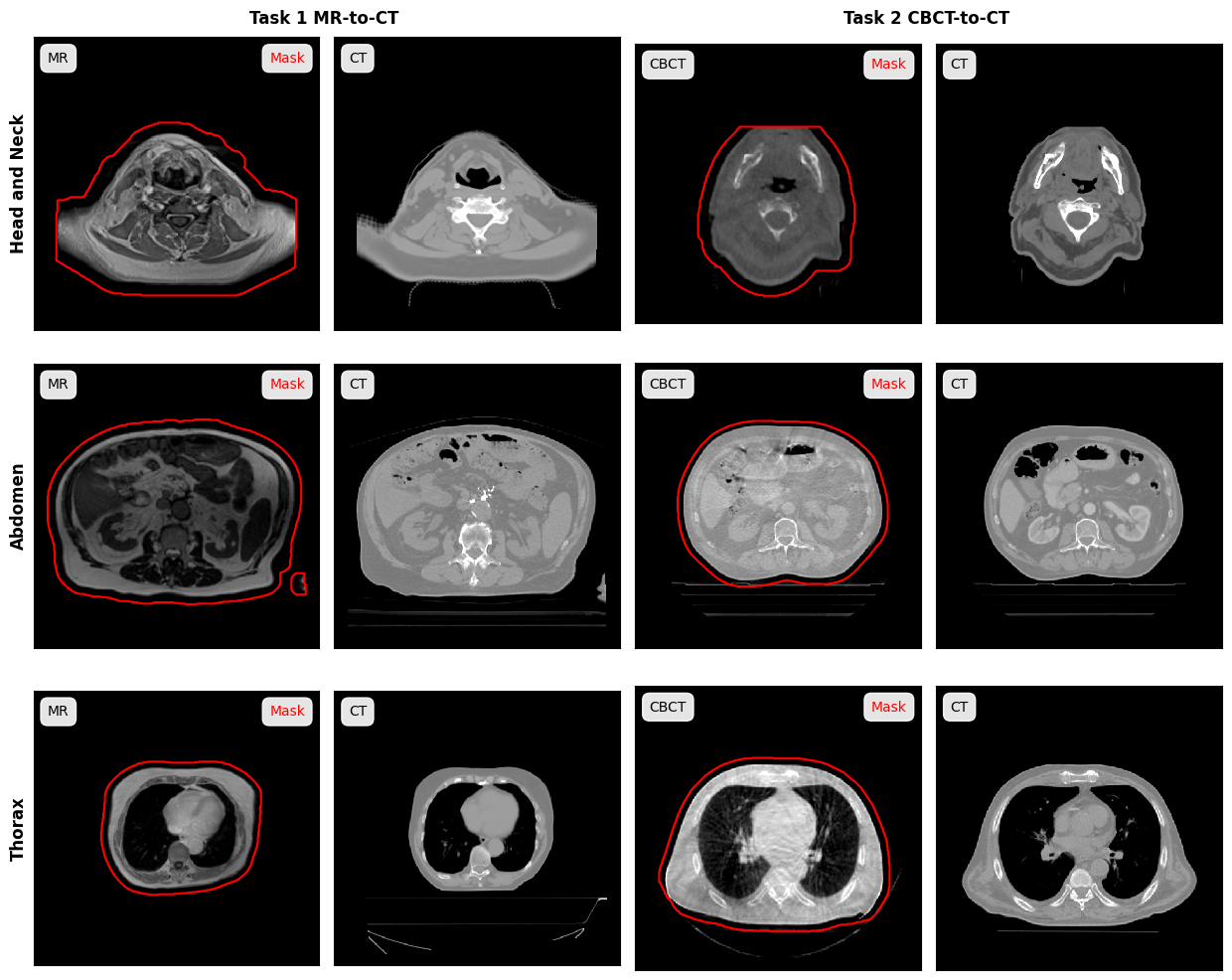
The figure below shows examples for each task and anatomical regions:
For each task and anatomy an overview folder is provided which contains the following files:
-
[task]_[anatomy]_parameters.xlsx: This file contains information about the image acquisition protocol for each patient.
-
[task][anatomy][Center][PatientID]_overview.png: For each patient a png showing axial, coronal and sagittal slices of CBCT/MR, CT, mask and the difference between CBCT/MR and CT is provided. These images are meant to provide a quick visual overview of the data.
Data License¶
The dataset is provided under two different licenses:
-
Data from centers A, B, C, and E is provided under a CC-BY-NC 4.0 International license (creativecommons.org/licenses /by-nc/4.0/).
-
Data from center D is provided with a limited license (Limited Use License Center D) that allows the use of the data solely for the challenge and is only valid as long as the challenge is active. By downloading the data, participants agree to these terms. Once the challenge ends, the data download will be deactivated and the data must be deleted.
Data Description¶
This challenge dataset contains imaging data of patients who underwent radiotherapy in the head and neck, thorax or abdomen region. Overall, the population is predominantly adult and no gender restrictions were considered during data collection. For Task 1, the inclusion criteria was the acquisition of a CT and MRI during treatment planning while for Task 2, acquisitions of a CT and CBCT, used for patient positioning, were required. Datasets for Task 1 and 2 do not necessarily contain the same patients, given the different image acquisitions for the different tasks.
Data was collected at five international centers:
-
Radboud University Medical Center, Nijmegen, The Netherlands
-
University Medical Center Utrecht, The Netherlands
-
University Medical Center Groningen, The Netherlands
-
University Hospital Cologne, Germany
-
LMU University Hospital Munich, Germany
For anonymization purposes, the provenience of the institution center is not provided, and each center is indicated with letters from A to E.
The following number of patients is available in the training, validation and test set:
Training¶
|
Head-and-Neck (HN) |
Thorax (TH) |
Abdomen (AB) |
|||||||||||||||
|
A |
B |
C |
D |
E |
Total |
A |
B |
C |
D |
E |
Total |
A |
B |
C |
D |
E |
Total |
Task 1 |
91 |
0 |
65 |
65 |
0 |
221 |
91 |
91 |
0 |
0 |
0 |
182 |
65 |
91 |
19 |
0 |
0 |
175 |
Task 2 |
65 |
65 |
65 |
65 |
65 |
325 |
65 |
65 |
63 |
63 |
65 |
321 |
64 |
65 |
62 |
53 |
65 |
309 |
Validation¶
|
Head-and-Neck (HN) |
Thorax (TH) |
Abdomen (AB) |
|||||||||||||||
|
A |
B |
C |
D |
E |
Total |
A |
B |
C |
D |
E |
Total |
A |
B |
C |
D |
E |
Total |
Task 1 |
14 |
0 |
10 |
10 |
0 |
34 |
14 |
14 |
0 |
0 |
0 |
28 |
10 |
14 |
3 |
0 |
0 |
27 |
Task 2 |
10 |
10 |
10 |
10 |
10 |
50 |
10 |
10 |
10 |
10 |
10 |
50 |
10 |
10 |
10 |
8 |
10 |
48 |
Testing¶
|
Head-and-Neck (HN) |
Thorax (TH) |
Abdomen (AB) |
|||||||||||||||
|
A |
B |
C |
D |
E |
Total |
A |
B |
C |
D |
E |
Total |
A |
B |
C |
D |
E |
Total |
Task 1 |
35 |
0 |
25 |
25 |
0 |
85 |
35 |
35 |
0 |
0 |
0 |
70 |
25 |
35 |
8 |
0 |
0 |
68 |
Task 2 |
25 |
25 |
25 |
25 |
25 |
125 |
25 |
25 |
25 |
24 |
25 |
124 |
25 |
25 |
25 |
20 |
25 |
120 |
In total, for all tasks and anatomies combined, 890 MRI-CT pair (578 training, 89 validation, 223 testing) for Task 1 and 1472 CBCT-CT pairs (955 training, 148 validation, 369 testing) are available in this dataset.
All images were acquired with the clinically used scanners and imaging protocols of the respective centers and reflect typical images found in clinical routine. As a result, imaging protocols and scanner can vary between patients. A detailed description of the imaging protocol for each image, can be found in spreadsheets that are part of the dataset release (see dataset structure).
Data was acquired with the following scanners:
- MRI: Philips Ingenia 1.5T/3.0T or Marlin 1.5 T, Siemens Avanto fit 1.5T or Siemens MAGNETOM Vida fit 3.0T, ViewRay MRidian;
- CBCT: Elekta XVI, IBA Proteus+, Varian OBI;
- CT: Philips Brilliance Big Bore, Siemens Biograph PET-CT, Siemens SOMATOM or Confidence or go.Open, GE Optima CT580, Siemens Confidence, Toshiba Aquilon/LB.
For Task 1 (MRI to CT) MRIs were acquired with a T1-weighted gradient echo or a balanced steady-state free-precession sequence. The exact acquisition parameters vary between patients and centers. For centers A, C and D, some MRIs were acquired with and some without Gadolinium contrast.
Pre-processing¶
The following pre-processing steps were performed on the data:
-
Conversion from dicom to mha
-
Rigid registration between CT and MR/CBCT
-
Anonymization (face removal, only for hand and neck patients)
-
Patient outline segmentation (provided as a binary mask)
-
Crop MR/CBCT, CT and mask to remove background and reduce file sizes
The code used to preprocess the images can be found at: https://github.com/SynthRAD2025/preprocessing.
